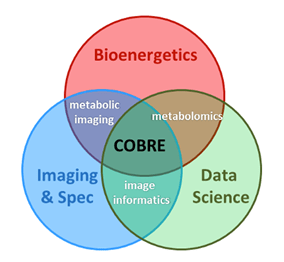
The AIMRC brings together a collection of resources and personnel to establish three Research Cores in Bioenergetics, Metabolic imaging and spectroscopy, and Data science/Big Data analytics, providing AIMRC members access to state-of-the-art metabolic research techniques and instruments.
Bioenergetics Core

Dr. Suresh Kumar Thallapuranam, Director
Dr. Thallapuranam is the Mildred-Cooper Endowed Chair of Biochemistry and Bioinformatics. He serves as Director of both the Protein and Peptide Synthesis Facility and NIH-funded NMR Facility on campus. For the past 15 years, Dr. Thallapuranam has been working on understanding the structure-function relationship and interactions of cytokines such as, the fibroblast growth factors, interleukin 1α, interleukin-12, and the vascular endothelial growth factor(s). Dr. Thallapuranam has over 20 years of working experience using techniques in the determination of 3D structures of proteins and protein complexes, such multidimensional NMR spectroscopy, X-ray crystallography, SAXS and in silico molecular modeling studies.
Initial capabilities of the Bioenergetics Core include:
- Live-cell metabolic flux assays through Agilent Seahorse XF analyzers
- Oroboros O2k-FluoRespirometer
Imaging and Spectroscopy Core

Dr. Narasimhan Rajaram, Director
Dr. Narasimhan Rajaram is an Associate Professor in the Department of Biomedical Engineering at the University of Arkansas, and serves as the Director of the Imaging and Spectroscopy Core. Dr. Rajaram currently directs the Laboratory for Functional Optical Imaging and Spectroscopy and is supported by NIH R01, NSF CAREER, and DoD funding mechanisms. Dr. Rajaram has 15 years of experience in the development of optical instrumentation and the use of optical microscopy and spectroscopy techniques for pre-clinical discovery work and clinical translation (>40 publications).

Dr. Timothy Muldoon, Associate Director
Dr. Timothy Muldoon, PhD, MD, is also an Associate Professor in the Department of Biomedical Engineering. Dr. Muldoon has 16 years of experience in the development of clinically compatible microendoscopes and advanced microscopic tools (>30 publications) and is funded by NIH and NSF CAREER awards.

Dr. Alan Woessner, Manager
Dr. Alan Woessner serves as the manager for the Imaging and Spectroscopy Core. Dr. Woessner was awarded a B.S. in Biomedical Engineering in 2016 and a Ph.D in Biomedical Engineering in 2022 from the University of Arkansas. He specializes in multiphoton microscopy, fluorescence lifetime imaging microscopy (FLIM), and image and data analysis.
The Imaging and Spectroscopy Core features the following equipment:
- Bruker Ultima Investigator upright multiphoton microscope
- Bruker Ultima Investigator inverted I multiphoton microscope
- Thorlabs inverted multiphoton microscope for 2D/3D cell culture and intravital microscopy
- Olympus Fluoview FV10i-LIV Confocal microscope
- JY Horiba Imaging spectrometer
- Andor Raman spectrometer
- Ocean Optics UV-Vis-NIR FLAME spectrometers
Initial capabilities of the Imaging and Spectroscopy Core include:
- Label-free multiphoton microscopy of NADH and FAD autofluorescence for in vivo or in vitro assessments of cell metabolism
- Diffuse optical spectroscopy for in vivo studies of tissue oxygenation
- Standard laser scanning confocal microscopy for live cell and tissue imaging
Data Science Core

Dr. Xintao Wu, Director
Dr. Xintao Wu, is a Professor of Data Science in the Department of Computer Science and Computer Engineering, and serves as director of the Data Science Core. His research interests include big data analysis, data mining, machine learning, data privacy, fraud detection, and fairness awareness learning. He has published >150 articles in peer-reviewed journals, is an associate editor and/or editorial board member of a number of prestigious Journals, and has served on NSF review panels and program committees as area chair, senior PC, and PC of top international conferences, including ACM KDD, WWW, CIKM, IEEE ICDM, SIAM SDM, AAAI, IJCAI, PKDD, and PAKDD. He also served as program co-chair of the 2020 IEEE International Conference on Big Data. (Published Works)
The AIMRC data science core offers dedicated personnel for one-on-one access to assist center members in analyzing and identifying relationships between large datasets. Although researchers often use bioinformatics to support analysis of large datasets, the AIMRC Data Science Core offers the unique advantage of using AI-based approaches.
Advantages of AI-based Approaches:
- Transformative impact on how image datasets are processed, analyzed, and quantified, and are enabling researchers to increase image resolution, identify image features, and predict clinical measurements and histology in ways not previously possible
- Extends beyond imaging applications! Deep learning provides new approaches to reconstructing metabolic pathways and analyzing metabolic flux without relying on the assumptions of traditional kinetic models
- Connect bioenergetics outcomes to larger data sets to predict disease severity and identify new therapeutic targets
Initial capabilities of the Data Science Core include:
- Development of custom data analytics for Project Leaders and Center members
- Computational time on GPU clusters and algorithm development services will be offered to aid with rapid neural training and algorithm development
The Data Science Core features the following major computational resources:
- Four Dell XE8545 dual CPU quad GPU servers with NVidia A100 GPUs, AMD 7543 CPUs, and 1024 GB of memory. They will be integrated with the other equipment of the AHPCC (Arkansas High Performance Computing Center), including four similar quad GPU servers each for DART (reference) and for UA Computer Science and Computer Engineering, and one quad GPU server for UA Biomedical Engineering
- 18 single A100 GPU dual CPU servers for DART
- Each A100 GPU delivers 9.7 Teraflops of double precision and up to 164 Teraflops of Tensor float performance. The system also includes 19 older single V100 GPU dual CPU servers and about 400 non-GPU dual GPU servers to produce in total more than one Petaflop of peak double precision performance
- Three Petabytes of DDN Lustre parallel storage
- Four PB of nearline Lustre and object storage
- Infiniband and Ethernet networks to act as a single system
- External interfaces such as login nodes, data transfer nodes, and 100 Gb high-speed ethernet to Internet2 and other educational and research networks
- Software resources include about 450 application packages installed from source or commercial sources, about 150 packages installed as python or R modules, and hundreds of packages installed as RPMs (system packages) from CentOS, EPEL, and OpenHPC. Many of these applications have several installed versions each
- Lmod allows a customized user environment with the software and versions selected from the many possibilities
- An OpenOnDemand portal, Jupyter Notebooks, RStudio, and other graphical interfaces are popular with new and educational users who aren’t accustomed to command line batch computing
- Globus endpoints provide easy data movement across systems and continents. Virtual machine and container capability allow complex and repeatable software installations
Training resources are vital to efficient use of a complicated instrument. AHPCC, UA IT Research Computing, and UA Libraries are supporting a local instance of the “Software Carpentry” training program with programs in python, git version control, and other topics. These are delivered state-wide along with custom courses in system usage and related topics