WELCOME
The Nakanishi Lab is located in the Department of Biological Sciences at the University of Arkansas, Fayetteville.
OUR CORE MOTIVATION
We aspire to understand where we come from – our deep evolutionary roots as animals – and how fundamental traits emerge in evolution.
WHAT WE DO
Our mission is to advance foundational knowledge in neurobiology, development and evolution through rigorous, question-driven science, while creating opportunities for diverse students to explore and experience the process of propelling scientific discoveries.
CURRENT RESEARCH INTERESTS

In the Precambrian ocean over hundreds of millions of years ago, the first animal evolved from a single-celled, flagellated ancestor. Critical to this transition was the emergence of development: the process by which a fertilized egg becomes organized into a multicellular body consisting of stably differentiated somatic cells and germ cells. Subsequently, animals diversified into 5 major lineages – Ctenophora (combjellies), Porifera (sponges), Placozoa, Cnidaria (jellyfishes, corals and sea anemones) and Bilateria (insects, worms and humans). Bilaterian radiation then generated chordates, from which evolved vertebrates, from which evolved mammals, primates, and then humans. Given the long history of evolution as animals, fundamental traits of humans (or any other animal) have their origins in the deep roots of the animal tree. Yet, when and how building blocks of basic human traits like hearing emerged in evolution remain unresolved. We work to address such questions.
In particular, we work on fundamental problems in neural development, function, and evolution by studying the biology of Cnidaria. We focus on investigating Cnidaria because of its deep phylogenetic history and diversity; it diverged from its sister group Bilateria (e.g. insects, worms and vertebrates) over 580 million years ago. As one of the early-evolving animal groups, Cnidaria has strong potential not only to provide us with information necessary to understand the biology of early animal ancestors, but also to enable the discovery of novel biology. Furthermore, a variety of experimental tools, including CRISPR-Cas9 genome editing, have been, and are being, established in Cnidaria, allowing mechanistic studies of cnidarian biology and evolution.
Research theme 1: Mechanisms of mechanoreceptor development and their evolution

In parallel with bilaterians, cnidarians have evolved morphologically diverse mechanosensory neural structures, ranging from hydroid filiform tentacles to jellyfish gravity-sensors (e.g. statocysts). Housed within the mechanoreceptors are the primary sensory neurons known as the concentric hair cells – characterized by an apical mechanosensor consisting of a non-motile cilium surrounded by one or multiple collars of microvilli/stereovilli. But it remains unclear whether deeply conserved or divergent genetics underlies the development of mechanosensory neurons/cells across Cnidaria and Bilateria. We are working to resolve the structure of the gene regulatory networks underpinning cnidarian mechanoreceptor development, using the sea anemone Nematostella vectensis as an experimental and comparative model.
Read Ozment et al. (2021) and Baranyk et al. (2025) to learn more.
Funding: NSF IOS 1931154, “Unveiling the evolution of neural differentiation mechanisms in animals: a study of the structure and function of the POU-IV/Brn-3 gene regulatory network in Cnidaria”; Arkansas Biosciences Institute.
Research theme 2: Neuropeptidergic control of life cycle transition

One of the unique and interesting aspects of cnidarian biology is their life cycle. Jellyfish, for example, undergo multiple rounds of life cycle transition that drastically alters form and behavior, and some even rejuvenate by reverse development. However, the mechanisms of metamorphic transformations are poorly understood.
Neuropeptides expressed in the larval nervous system are thought to regulate the transition into a sedentary polyp (sea anemone-like) form, and we have previously shown that deeply conserved neuropeptides accelerate life cycle transition in the sea anemone Nematostella vectensis. We are currently investigating the mechanism by which this occurs. More specifically, we aim to identify 1) the neuropeptide receptors and 2) immediate response genes of neuropeptidergic signaling.
Read Nakanishi and Martindale (2018), Nakanishi and Jacobs (2019), and Zang and Nakanishi (2020) to learn more.
Funding: NSF IOS 2042529, “CAREER: Neuropeptidergic control of life cycle transition in Cnidaria”.
JOIN THE LAB
We welcome applications from prospective postdocs and students (at graduate and undergraduate levels) – see OPPORTUNITIES below for details. Research and teaching assistantships are available for graduate students. Interested applicants should send me an email (at nnakanis@uark.edu) containing a brief summary of your qualification, research interests and goals, along with your CV.
OUR CNIDARIAN MODELS
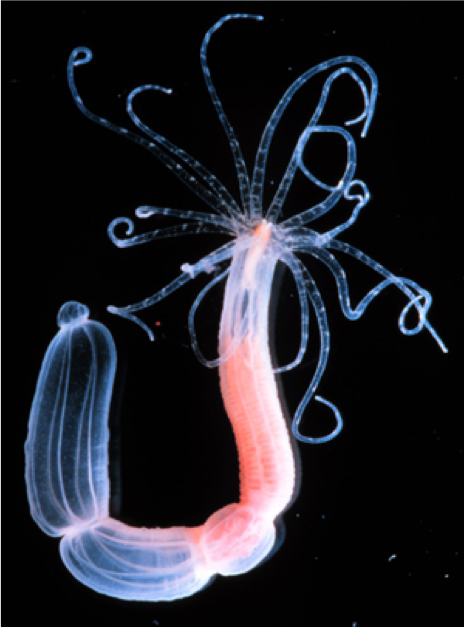
Anthozoan cnidarian Nematostella vectensis (starlet sea anemone)

Scyphozoan cnidarian Aurelia (moon jelly)
SELECT PUBLICATIONS (see Google Scholar for a complete list)
Leach, W. B., Babonis, L., Juliano, C. E., Nakanishi, N., Schnitzler, C. E., Steinmetz, P. R. H., and Layden, M. J. 2025. Discoveries and innovations in cnidarian biology at Cnidofest 2024. EvoDevo. 16:9. https://doi.org/10.1186/s13227-025-00247-5
Nakanishi, N., Takahashi, M., and Kumano, G. 2025. Diversification of cnidarian mechanosensory neurons across life cycle phases: evidence from Hydrozoa. Integrative and Comparative Biology, icaf027, https://doi.org/10.1093/icb/icaf027.
Baranyk, J., Malir, K., Silva, M. A. P., Rieck, S., Scheve, G., and Nakanishi, N. 2025. Structural molecular and developmental evidence for cell-type diversity in cnidarian mechanosensory neurons. Nature Communications 16:1514. https://doi.org/10.1038/s41467-025-56115-2
Ozment, E., Tamvacakis, A. N., Zhou, J., Rosiles-Loeza, P. Y., Escobar-Hernandez, E. E., Fernandez-Valverde, S. L., and Nakanishi, N. 2021. Cnidarian hair cell development illuminates an ancient role for the class IV POU transcription factor in defining mechanoreceptor identity. eLife 2021;10:e74336 DOI: 10.7554/eLife.74336
Zang, H. and Nakanishi, N. 2020. Expression analysis of cnidarian-specific neuropeptides in a sea anemone unveils an apical-organ-associated nerve net that disintegrates at metamorphosis. Front. Endocrinol. 11:63. doi: 10.3389/fendo.2020.00063
Nakanishi, N. and Jacobs, D.K. 2020. The Early Evolution of Cellular Reprogramming in Animals. In Evolutionary Cell Biology, Deferring Development: Setting aside cells for future use in development and evolution. Brian K. Hall and Corey Bishop. Eds. CRC press.
Silva, M.A. and Nakanishi, N. 2019. Genotyping of sea anemone during early development. J. Vis. Exp. (147), e59541, doi:10.3791/59541.
Nakanishi, N. and Martindale, M.Q. 2018. CRISPR knockouts reveal an endogenous role for ancient neuropeptides in regulating developmental timing in a sea anemone. eLife 2018;7:e39742 DOI: 10.7554/eLife.39742.
Nakanishi, N., Camara, A., Yuan, D.C., Gold, D.A. and Jacobs, D.K. 2015. Gene expression data from the moon jelly, Aurelia, provide insights into the evolution of the combinatorial code controlling animal sense organ development. PLoS ONE 10(7):e0132544.
Nakanishi, N., Renfer, E., Technau, U. and Rentzsch, F. 2012. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development. 139(2):347-357 Faculty of 1000 article (rated Ffa6): 2012. F1000.com/1349198
Nakanishi, N., Yuan, D., Hartenstein, V. and Jacobs, D.K. 2010. Evolutionary origin of rhopalia: insights from cellular-level analyses of Otx and POU expression patterns in the developing rhopalial nervous system. Evolution and Development. 12(4):404-415.
Nakanishi, N., Yuan, D., Jacobs, D.K., and Hartenstein, V. 2008. Early development, pattern and reorganization of the planula nervous system in Aurelia (Cnidaria, Scyphozoa). Development Genes and Evolution. 218(10):511-524.
CURRENT MEMBERS

Nagayasu Nakanishi
Principal investigator / Associate Professor
2021 Robert C. and Sandra Connor Endowed Faculty Fellow
2021 NSF CAREER award recipient

Ndotimi Apulu

Julia Baranyk
Ph.D student, Doctoral Academy Fellow
2025 David Causey Prize for Graduate Study recipient

Emily Ye
Undergraduate honors student – Junior

Paige Norris
Undergraduate honors student – Junior
Kailey Hall
Lab manager/research associate
PAST MEMBERS
Postdoctoral fellows
Dr. Arianna Tamvacakis, 2019-2021
Graduate students
Aparajita Das, 2023-2025. Aparajita has successfully defended her MSc thesis and will graduate in Spring, 2026.
Caroline Johnson, 2024-2025.
Miguel Silva, 2019-2023. Miguel graduated with an MSc degree. Miguel currently pursues a PhD at the Institute of Biophysics and Biomedical Engineering, Faculty of Sciences, University of Lisbon, in Portugal.
Ethan Ozment, 2020-2022. Ethan currently works as an electron microscopy specialist at UT Southwestern Medical Center.
Undergraduate students
Sakura Rieck (Univ. of Arkansas), 2022 Fall – 2024 Fall. Sakura received the Student Undergraduate Research Fellowship in 2024.
Emily Bullock (Univ. of Arkansas), 2023 Spring – 2024 Spring. Emily graduated with Cum Laude and will be pursuing an MSc in Physiology at Georgetown University.
Rishi Parvathanenni (Univ. of Arkansas), 2023 Spring – 2024 Spring.
Kathryn Wootten (Univ. of Arkansas), 2023 Spring – 2024 Spring
Gracie Scheve (Bowdoin College), 2023 Fall. Gracie has been nominated by Bowdoin College for the Barry Goldwater Scholarship. Gracie is set to pursue a PhD in William Browne lab from Fall 2025.
Kristen Malir (Univ. of Arkansas), 2020 Fall – 2022 Spring. Kristen graduated with Magna Cum Laude. She currently works at the Cytogenetics Laboratory of the University of Kansas Medical Center.
Julia Baranyk (Univ. of Arkansas), 2020 Spring – 2021 Spring. Julia graduated with Cum Laude and started pursuing an MSc degree in the N lab from Fall 2021 and PhD degree from Fall 2024.
Allison Shildt (Univ. of Arkansas), 2019 Fall- 2021 Spring. Allison graduated with Summa Cum Laude, and started attending the Saint Louis University School of Medicine from Fall 2021.
Katherine McTigrit (Univ. of Arkansas), 2018 Fall -2021 Spring. Katie graduated with Cum Laude, and began attending the University of Arkansas for Medical Sciences from Fall 2021.
Hannah Zang (Lyon College), 2019 Summer, INBRE research program; her Frontiers in Endocrinology publication was featured in Lyon College News on February 27, 2020. In March 2020, Hannah was selected as a 2020 Barry Goldwater Scholar. She started attending the Duke University School of Medicine from Fall 2021.
Delaney Cross (Univ. of Arkansas), 2018 Fall -2019 Spring
High School student volunteers
Sakura Rieck (Fayetteville High School). Sakura began attending the University of Arkansas from Fall 2021, and returned to the N lab as an honors student from Fall 2022.
Research associates
Jianhong Zhou, 2018-2022
Sai Divya Challapalli, 2018
OPPORTUNITIES
Undergraduate student position:
I am interested in having one or two highly motivated undergraduate researchers from each level (typically Sophomore, Junior and Senior). If you would like to join us to do research in neuro-evo-devo, contact me at nnakanis@uark.edu with your CV, transcript, and a brief description of why you want to join the lab. Preference will be given to students who have successfully completed Genetics (BIOL 2323).
Graduate student position:
The Nakanishi lab at the University of Arkansas seeks applicants interested in pursuing an MSc or PhD in biology to start in the Fall of 2026. The lab is interested in addressing fundamental questions in neural development, function, and evolution (“neuro-evo-devo”) by studying the biology of early-diverging animal groups – cnidarians (sea anemone and jellyfish) in particular. Current NSF-funded research projects in the lab use the sea anemone Nematostella vectensis to investigate 1) the mechanism by which a single transcription factor controls distinct sets of target genes in different neural cell types, and 2) the mechanism by which neuropeptides modulate the timing of life cycle transition. Students may work on these and related problems in neuro-evo-devo. Student’s specific project will be developed based on the student’s research interests and educational background, in consultation with the PI of the lab. Research projects may involve gene expression analyses (e.g. in situ hybridization and immunohistochemistry), reverse genetics (e.g. CRISPR-Cas9), embryology (e.g. descriptive morphology, cell-lineage tracing and tissue transplantation), genomics (e.g. RNA-seq and ChIP-seq), and/or advanced microscopy (confocal and electron microscopy, and live-cell imaging). Research and teaching assistantships are available.
Requirements: Bachelor’s degree in biology or related field. An ideal candidate will have successfully completed undergraduate coursework in evolutionary biology and genetics. Research experiences in molecular biology, developmental biology, neurobiology, genomics/bioinformatics and/or microscopy techniques are preferred. Knowledge of invertebrate zoology is a plus but not required.
Application: The applicant may apply through the Department of Biological Sciences or through the Cell and Molecular Biology (CEMB) program. The deadline for Fall admission to the Biology Department is January 15, 2026, and that to CEMB January 1, 2026. In order to apply, the applicant will need to submit 1) an official graduate school application form, 2) official academic transcripts, 3) GRE scores, 4) three letters of recommendation from referees in academia, and 5) a program-specific application form (for CEMB). In addition, the applicant requires a faculty sponsor for admission; the applicant should send an email to Nagayasu Nakanishi (nnakanis@uark.edu) before applying. This email should contain a brief summary of your qualification, research interests and goals, your CV, copies of your academic transcripts, and GRE scores (if available).
Postdoc position:
An NSF-funded postdoctoral fellow position is available in the Nakanishi lab at the Department of Biological Sciences, University of Arkansas. The primary field of the lab’s research is cnidarian evolutionary developmental biology. In particular, this NSF-funded project will investigate the mechanism of life cycle transition, focusing on elucidating how neuropeptides control this process using the sea anemone Nematostella vectensis.
We are seeking a highly motivated and independent scientist with strong interests in advancing our understanding of the mechanism of cnidarian life cycle transition. The specific focus of the project will include 1) identification of neuropeptide receptors that are involved in metamorphosis regulation, and 2) characterization of the identity and function of transcription factors that are regulated by neuropeptidergic signaling at metamorphosis. In addition, the postdoc will be encouraged to develop an independent line of research that is broadly related to the NSF project theme and research interests of the lab. Moreover, the postdoc will have opportunities to mentor diverse students at the high school, undergraduate and graduate levels.
This is a full time, 40 hour per week position. The salary is commensurate with experience and education and includes full benefits. This is a one-year appointment, with the possibility to extend up to two additional years renewable based on the need for the position, availability of funding, and continued satisfactory level of performance in the role.
To be considered for this position, please send a curriculum vitae, a cover letter/letter of application, and a list of three professional references (name, title, email address, and contact number) to Nagayasu Nakanishi at nnakanis@uark.edu.
FUNDING SOURCES





